Client¶
Preamble¶
In order for the quality control to work optimally, it is recommended to split multiallelic variants. To compare similar variants during the Association Tests, input VCF files need to be annotated with vep (to added the consequence on genes and GnomAD frequencies). The command line to do this (after having installed vep) is:
/path/to/vep --cache --merged --offline --dir [/path/to/cache] --fork 24 --buffer_size 25000 --species homo_sapiens --assembly GRCh37 --use_given_ref --check_existing --allele_number --symbol --af_gnomad --vcf -i input.vcf.gz -o annotated.vcf
Launch the Client GUI¶
java -jar PrivAS.Client.jar [/path/to/data/directory]
Performing Association Tests with PrivAS¶
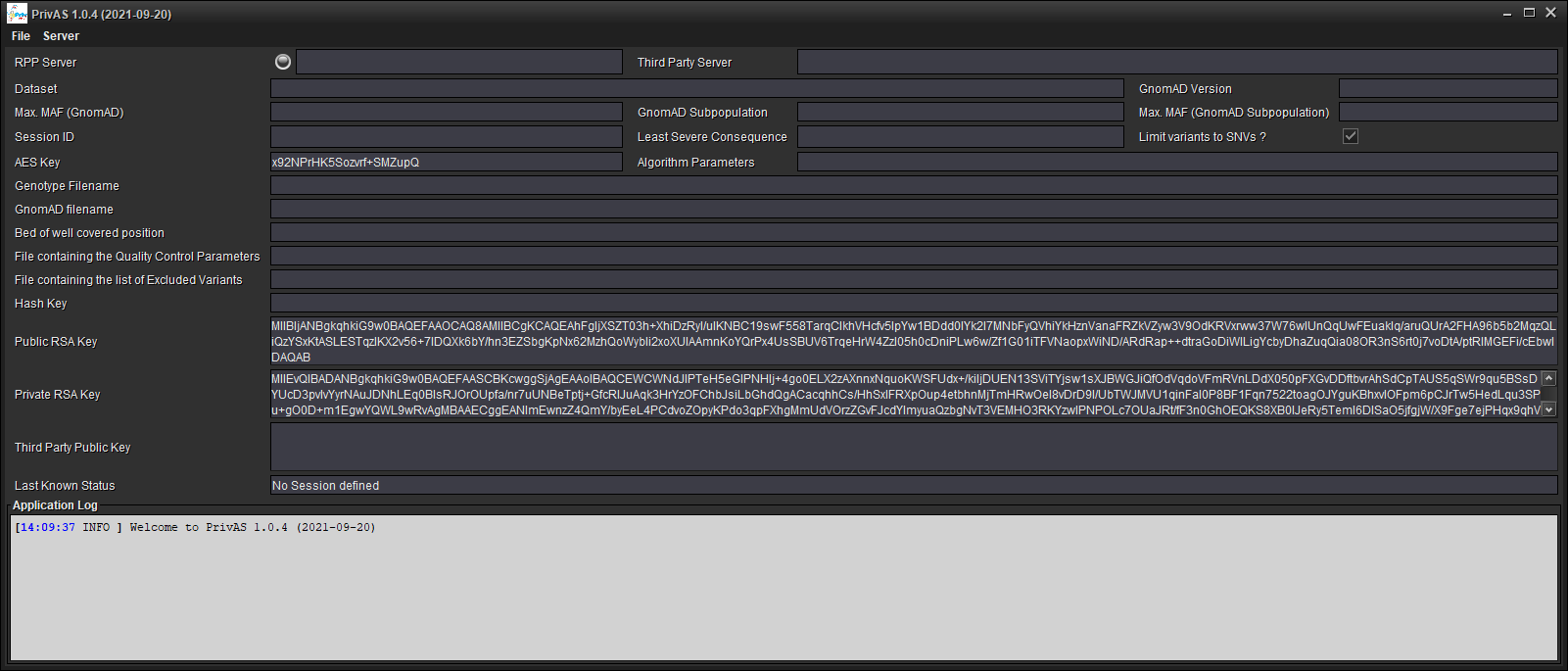
1. Load Variants¶
Load your annotated VCF file and perform a Quality Control on the Variants and Annotate with GnomAD frequencies [File/Load VCF, Apply QC and Annotate...]. Alternatively if you have already performed this operation for a previous session and you want the same QC parameters, you can simple load the results genotypes file, this will be much quicker [File/Load Annotated QCed Genotypes...]
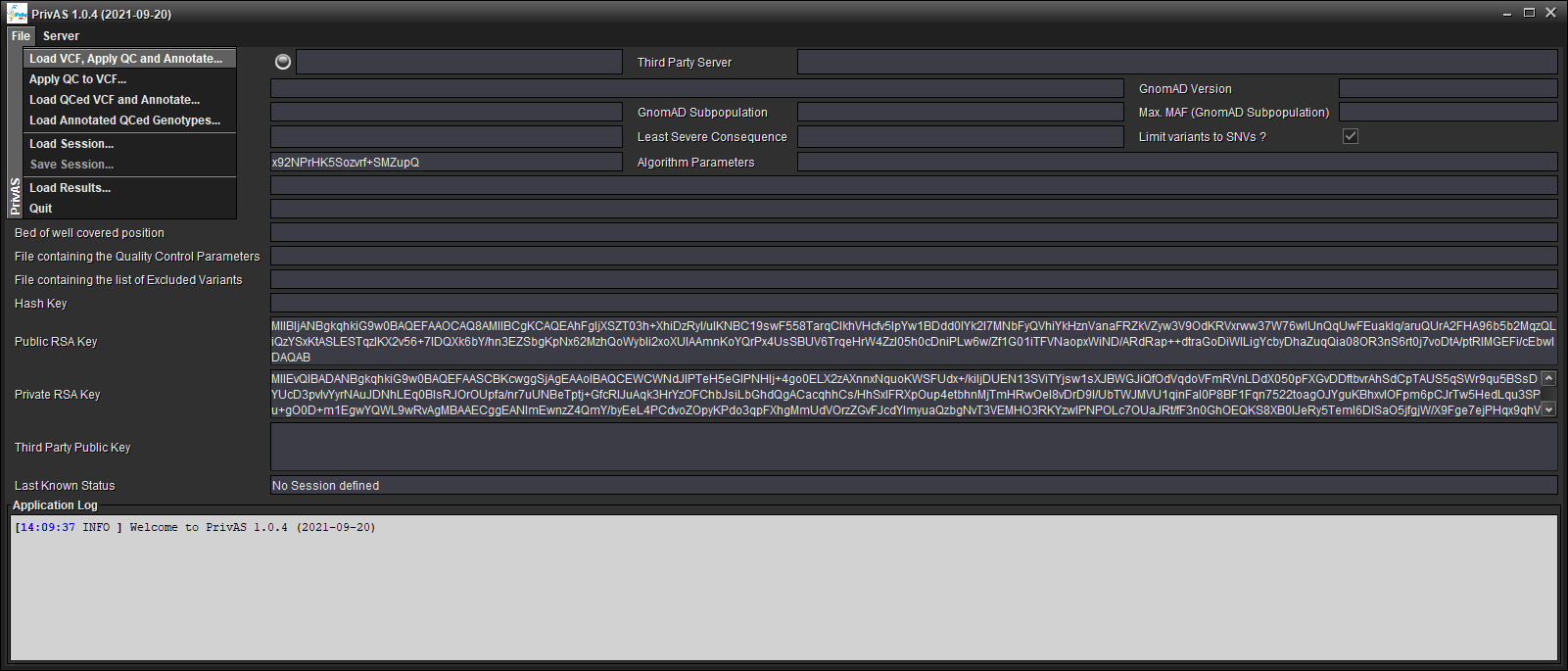
Choose your input VCF file
Choose a previous QC Parameters file…
Choose a GnomAD File
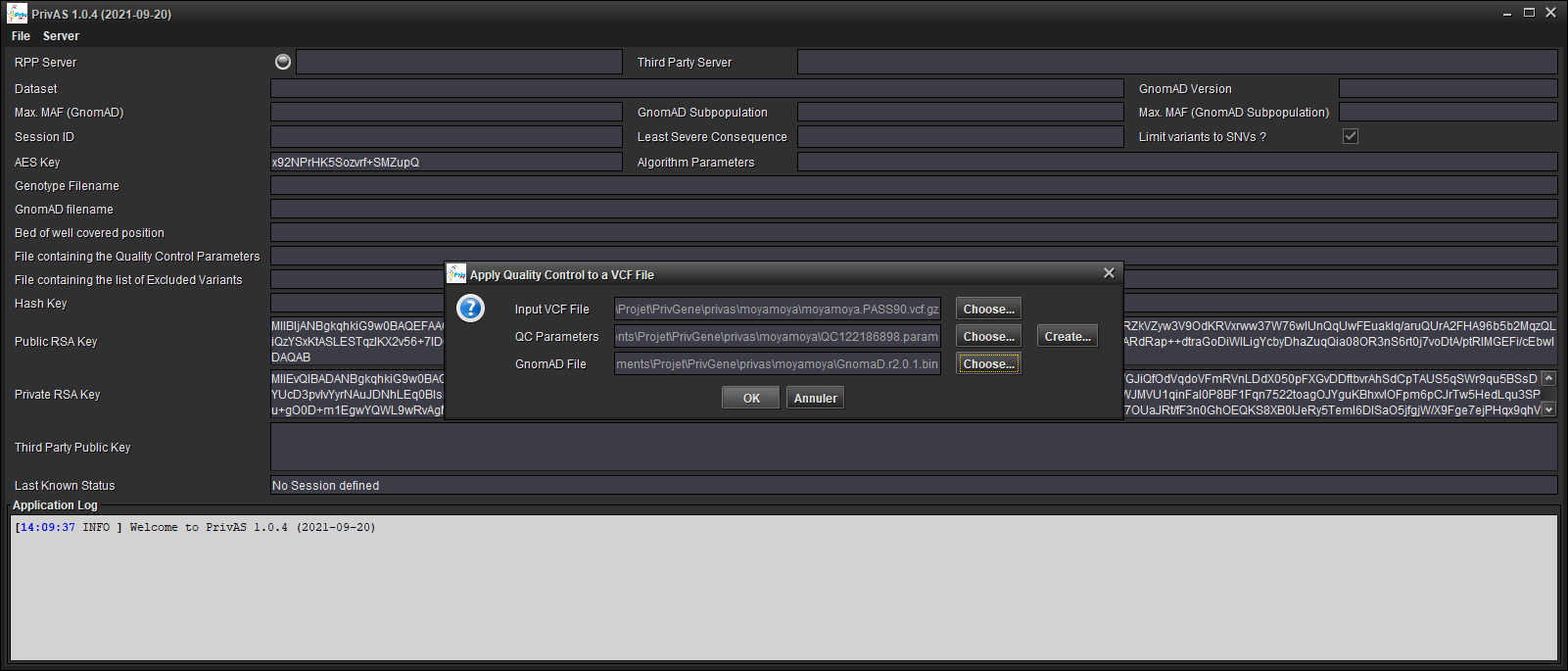
…or create a new QC Parameters file
The variants are Loaded (as seen in the [Genotype Filename] field)
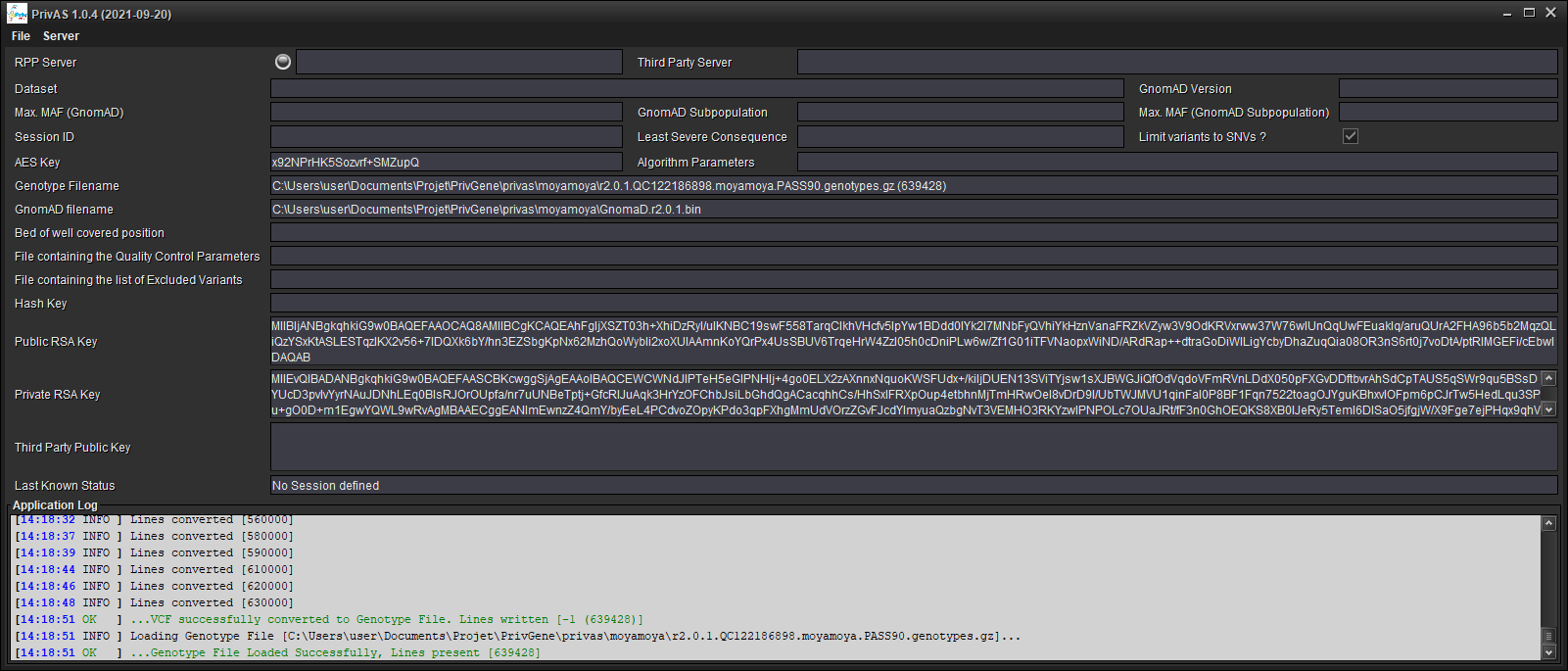
2. Connect to a RPP Server¶
Connect to a server [Server/Connect to RPP Server...]
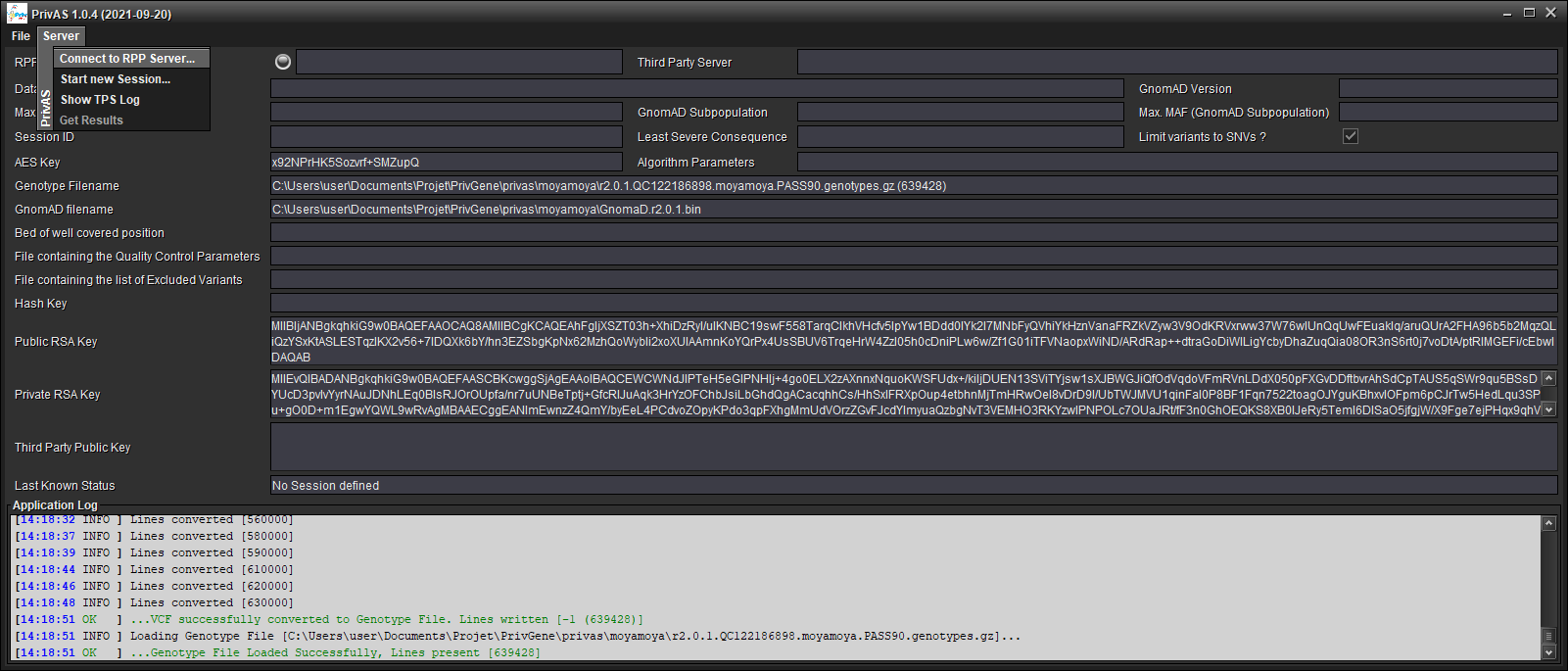
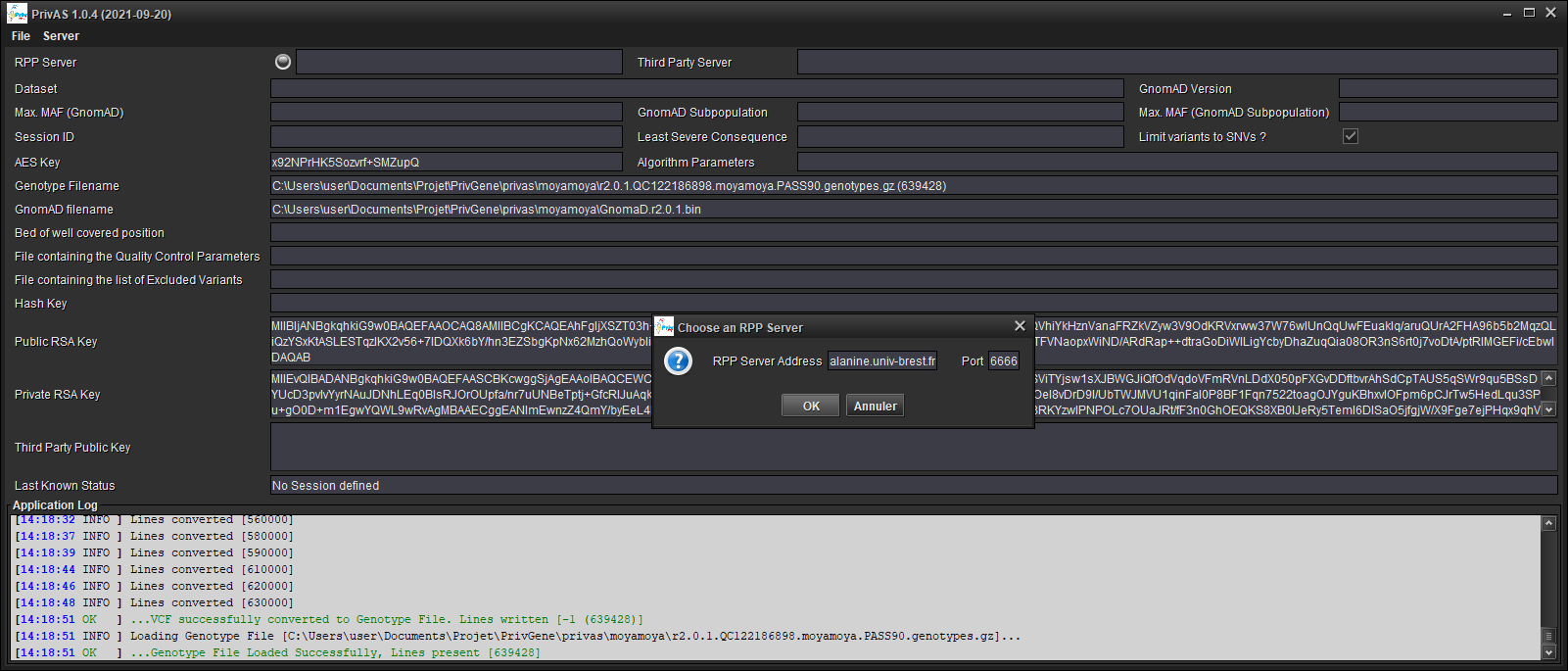
Fill in the address
Fill in the port number (default values point to a Server providing the FrEx datasets)
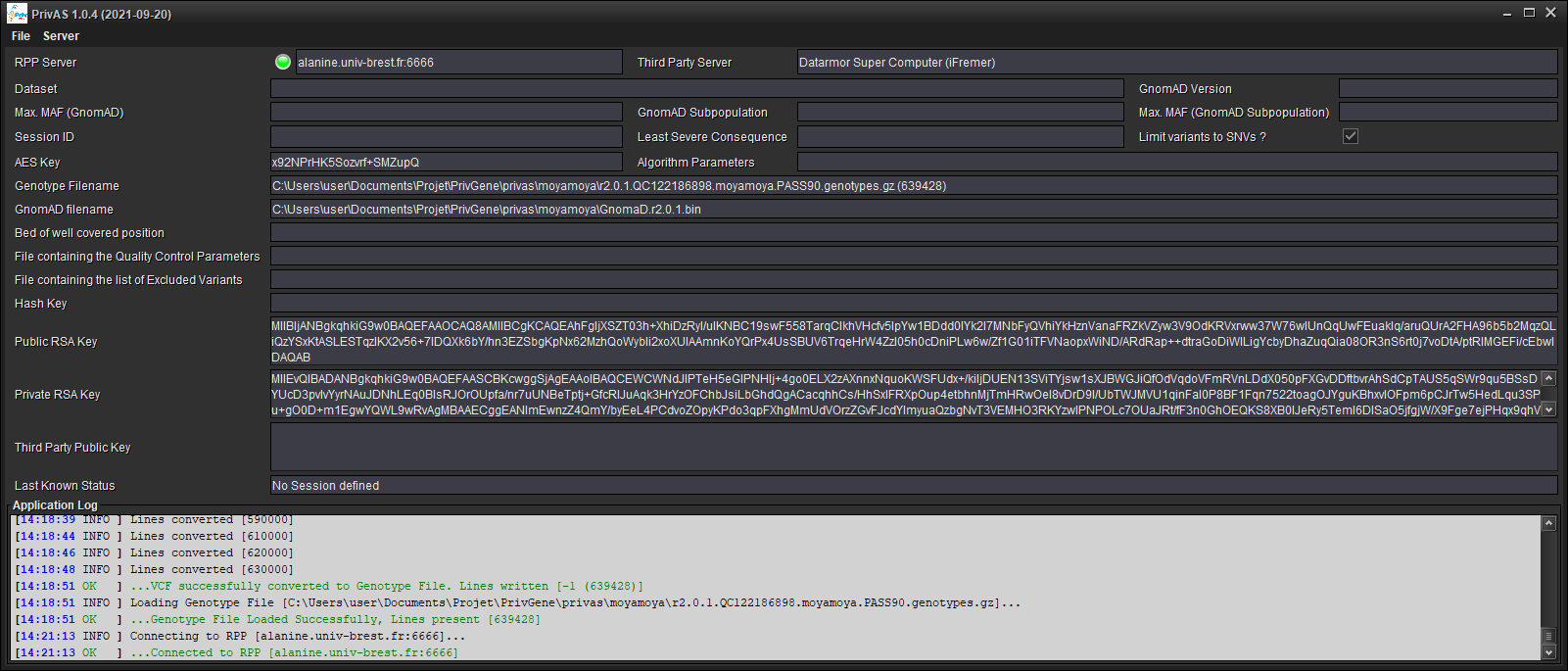
You are connected (as seen in the [RPP Server] field)
3. Perform Association Tests¶
Start a Session : [Server/Start new Session...]
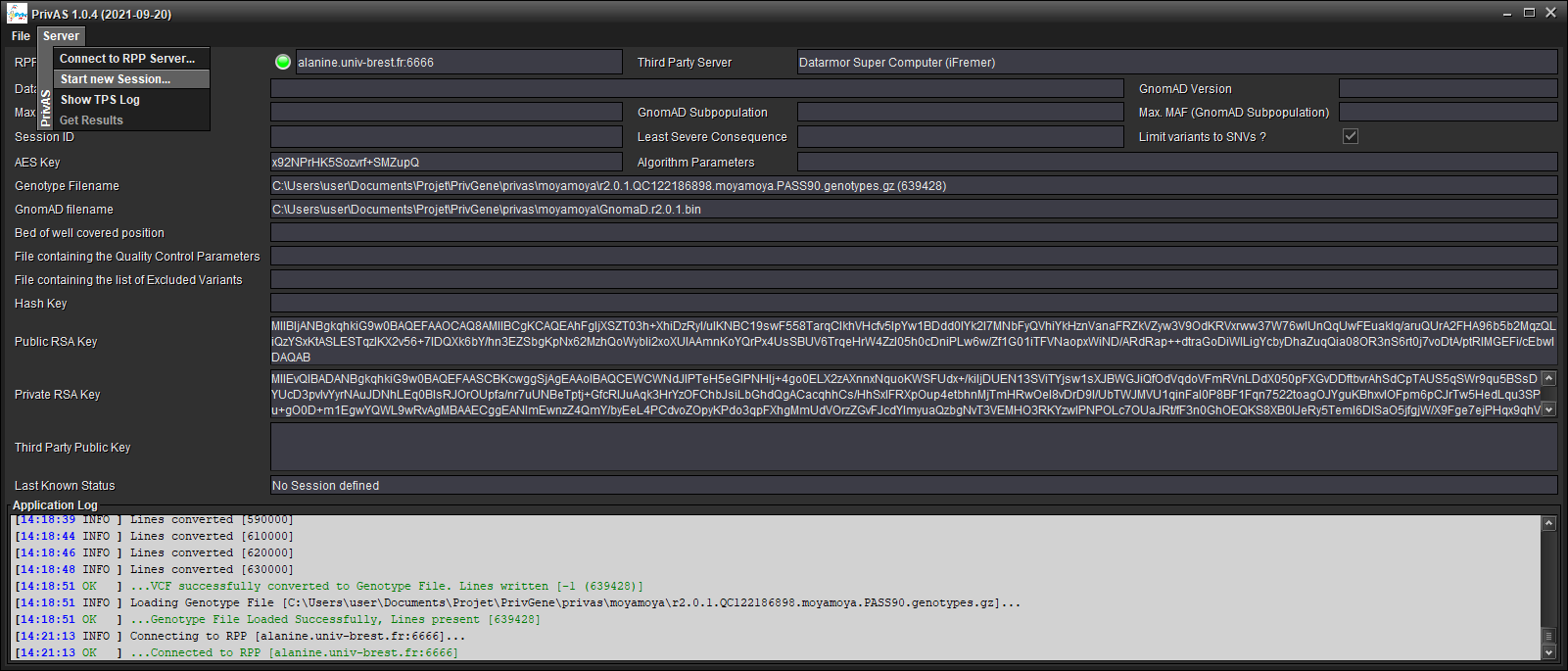
Choose:
one of the provided datasets (your data and RPP’s data must share the same Reference Genome)
the same GnomAD Version as the one you annotated your data with
the least severe VEP Consequence
the maximal frequency in GnomAD
a GnomAD subpopulation (or
Noneto disable)the maximal frequency in the GnomAD subpopulation
if you want to limit the tests to SNVs (as a INDEL calling is often less reliable)
the Bed file defining the well covered positions (any variants found outside those regions will be ignored)
the QC Parameters file used to extract the Client’s variants, so that the RPP’s variants will be filtered using the same criteria (should be automatically filled in)
the file listing the variants excluded by the QC (should be automatically filled in)
the Association Tests Algorithm (only WSS is present at this time) and its parameters
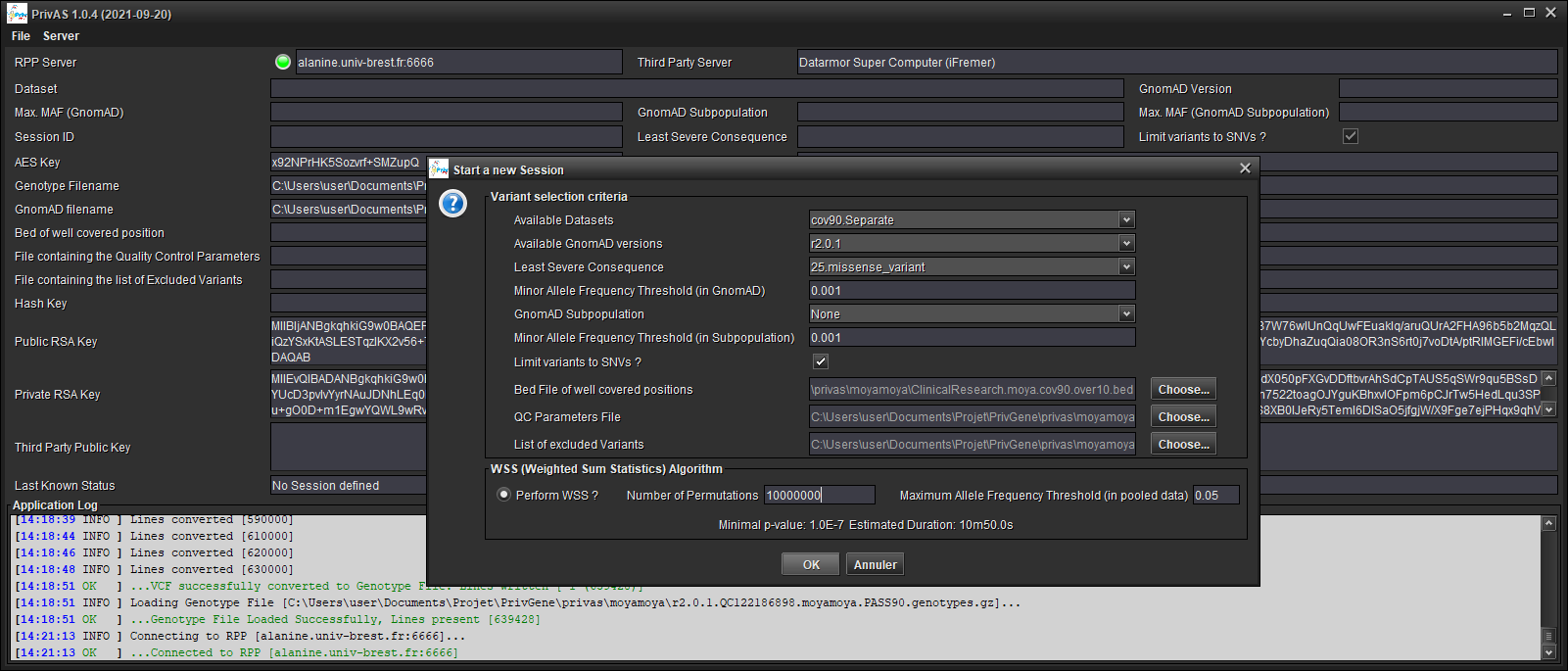
When prompted, save your session (this will allows you to keep track of the parameters you have selected, and to reconnected to the RPP if you have been disconnected).
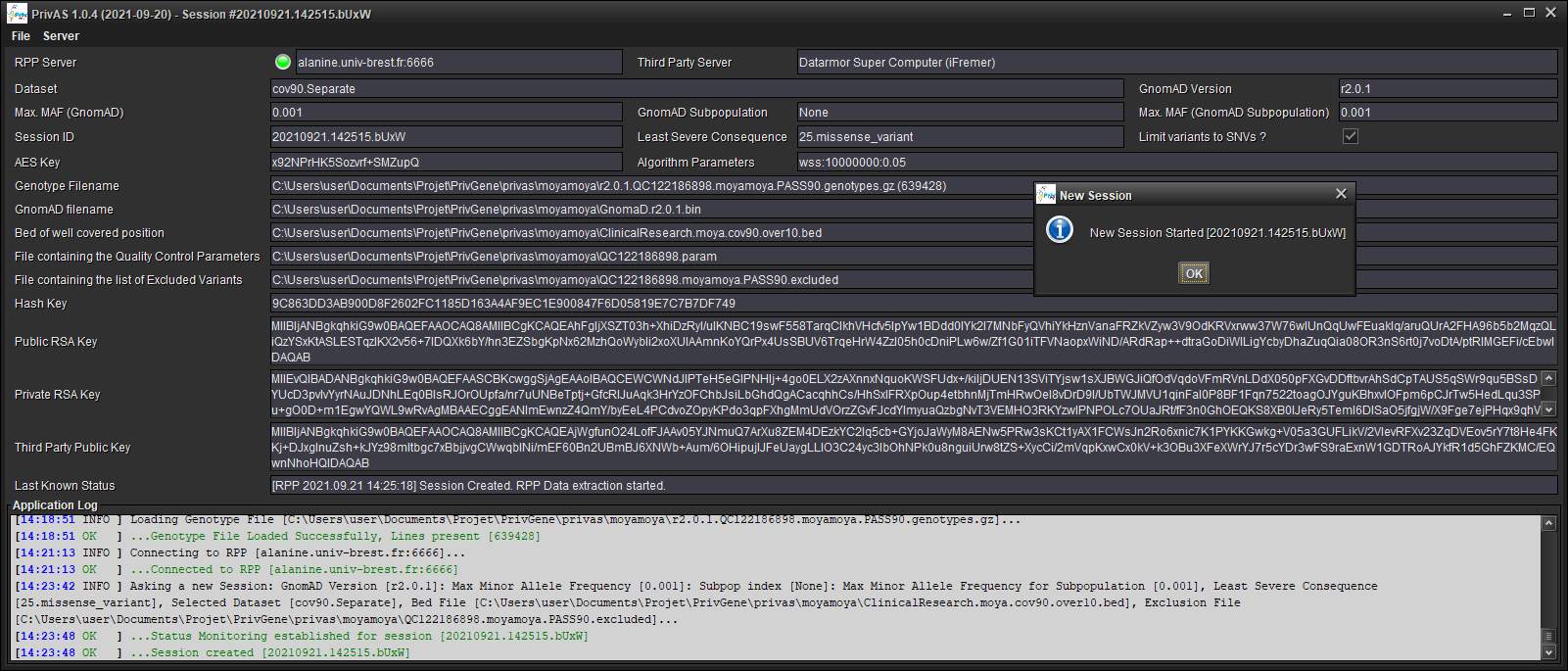
4. Follow your Association Tests progress¶
Follow the progress of the session in the [Last Known Status] Bar for the RPP server and in the [Third Party Server Log] window
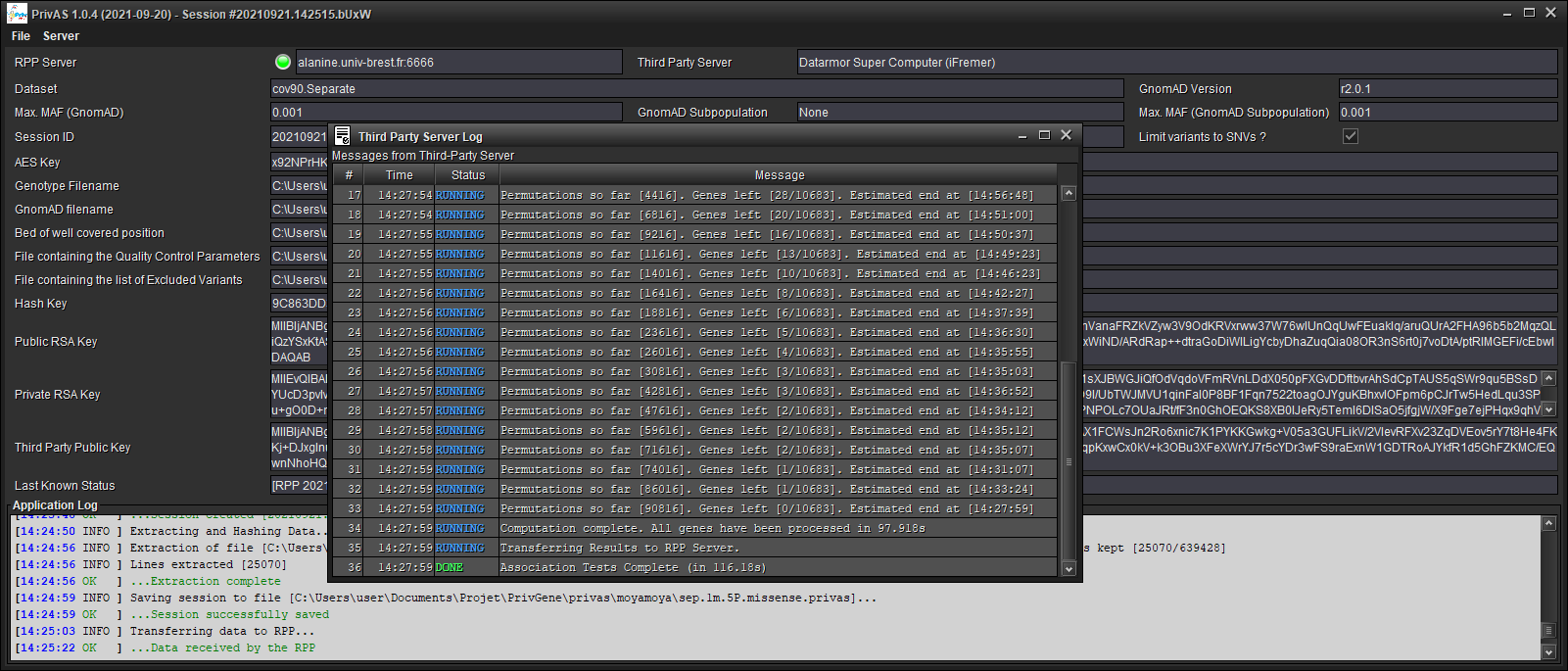
Save your results (when prompted)
5. Visualize your results¶
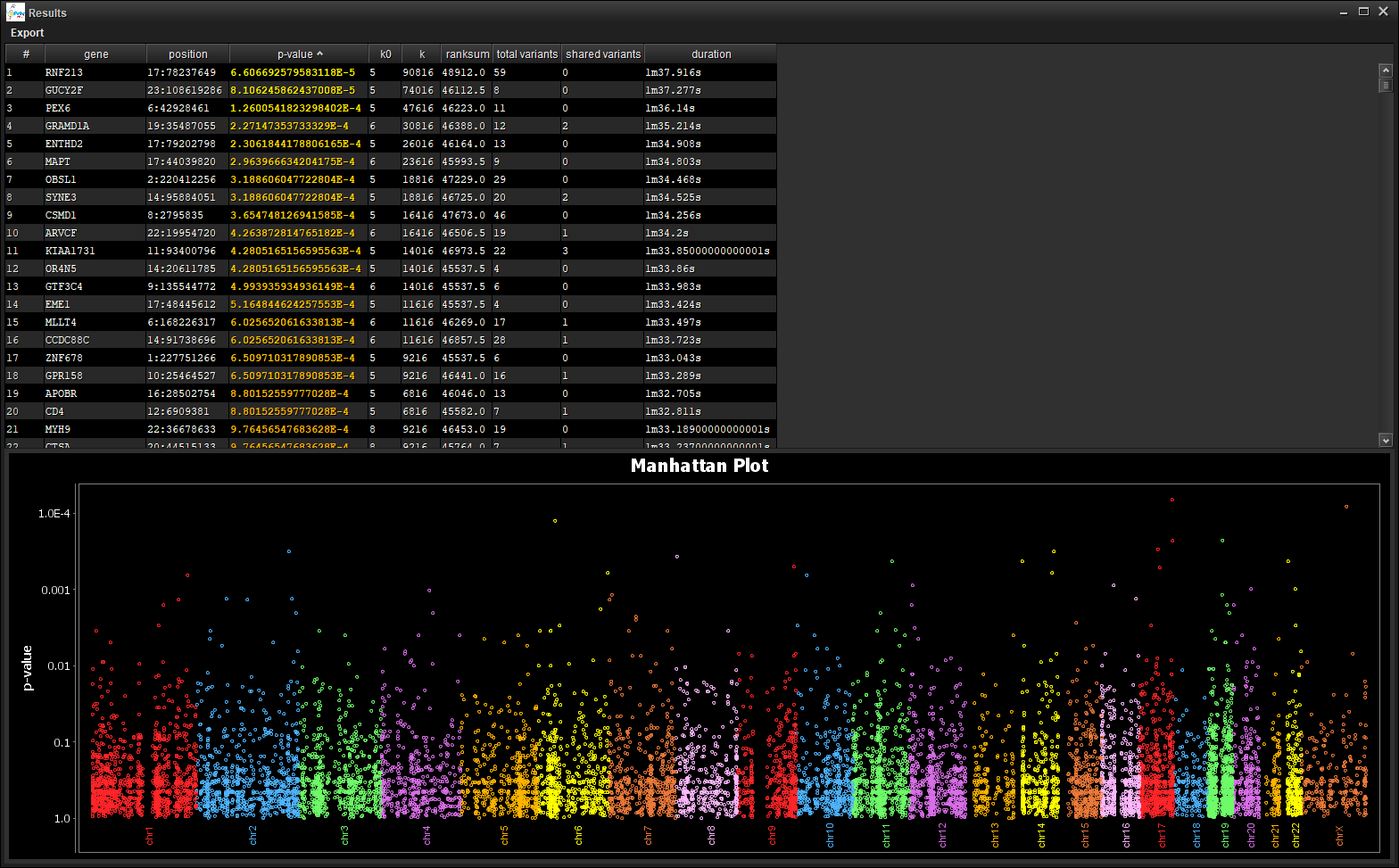
Save the the visualization (Table / Manhattan Plot) through the [Export] menu
The Main Window¶
Description of the various information found in the Main Window
Field |
Description |
|---|---|
RPP Server |
Reference Panel Provider (RPP) Server The address and port of the RPP Server. The Color indicates the state of the server :
|
Third Party Server |
Third Party Server (TPS) Name of the Server that will perform the actual calculations |
Dataset |
Name of the reference Dataset |
GnomAD Version |
Version of the GnomAD Version on the Reference Panel Provider |
Max. MAF (GnomAD) |
Maximum Minor Allele Frequency Threshold When selecting variants, the Client and the Reference Panel Provider will only keep variants with MAF below or equal to this threshold. |
Max. MAF (GnomAD Subpopulation) |
Same as above, but for frequencies in Selected Subpopulation |
GnomAD Subpopulation |
Selected subpopulation on GnomAD |
Session ID |
Session ID Uniquely identifies your work session for
|
Least Severe Consequence |
Least Severe Consequence When selecting variants, the Client and the Reference Panel Provider will only keep variants with Consequence above or equal to this threshold. |
AES Key |
AES Key Shared between
Data exchanged are encrypted/decrypted using this key. Thus the Reference Panel Provider (that serves as a bridge) cannot read these data. |
Limit variants to SNVs ? |
Limit To SNVs ? When selecting variants, the Client and the Reference Panel Provider will only keep variants that are SNVs. |
Algorithm Parameters |
Algorithm Parameters The algorithm and parameters that will be used by the Third-Party Server. |
Genotype Filename |
The Genotype File that was/will be used to extract/hash the data matching selection criteria. |
GnomAD filename |
GnomAD binary file |
Bed of well covered position |
Bed file of well covered positions |
Hash Key |
Hash Key Shared between
This key will be used to hash gene names and variant information, so that the Third-Party Server can do comparison and computing while not being able the read data either from the Client or the Reference Panel Provider. |
Public RSA Key |
Public RSA Key the Reference Panel Provider uses this key to encrypt the Hash Key, so that it is not legible on the network |
Private RSA Key |
Private RSA Key the Client use this key to decrypt the Hash Key. Only the Client know this key. |
Third Party Public Key |
Third Party Public Key This key is used to encrypt your AES key and share it with the Third-Party Server. This encryption prevents the Reference Panel Provider from reading the AES key. |
Last Known Status |
Last Known Status The Last known message sent by the Reference Panel Provider. |
Application Log |
Session Log.
|
PrivAS Client’s Command Lines¶
Main Command¶
Launch the Client GUI¶
java -jar PrivAS.Client.jar [Directory]
[Directory] |
initial working directory (Optional, default is current directory) |
Tools¶
Perform DEFAULT Quality Control on a VEP Annotated VCF file and convert the result to genotype file¶
java -jar PrivAS.Client.jar vcf2genotypes input.vcf(.gz) GnomADFile.bin
input.vcf(.gz) |
The Input VCF file (must have been annotated with vep) |
GnomADFile.bin |
The GnomAD file to use for the frequency annotation |
Perform a Quality Control on a VCF file¶
java -jar PrivAS.Client.jar vcf2qc input.vcf(.gz) qc.param
input.vcf(.gz) |
The Input VCF file (must have been annotated with vep) |
qc.param |
The file containing the QC parameters to apply |
Convert a QCed VEP Annotated VCF file to a genotypes file¶
java -jar PrivAS.Client.jar qc2genotypes vep_annotated_QCed_file.vcf(.gz) GnomADFile.bin
vep_annotated_QCed_file.vcf(.gz) |
The input VCF File (must result from a PrivAS QC and thus is annotated with vep) |
GnomADFile.bin |
The GnomAD file to use for the frequency annotation |
Creates an annotation binary file from lists of GnomAD (exome/genome) VCF files¶
java -jar PrivAS.Client.jar gnomad gnomADVersion listExomeVCFFiles.list listGenomeVCFFiles.list output.bin
gnomADVersion |
The name of the GnomAD Version |
listExomeVCFFiles.list |
File listing input GnomAD Exome files (one path per line) |
listGenomeVCFFiles.list |
File listing input GnomAD Genome files (one path per line) |
output.bin |
The name of the resulting binary file |