PrivAS¶
PrivAS is a tool to perform Association Tests on rare variants in a Privacy Preserving fashion.
It relies on a distributed solution, where two parties (a Client with genetic data from patients, and a Reference Panel Provider (RPP) with genetic data from healthy individuals) pool their data together without sharing them with each other. Instead, several encryption mechanisms (RSA, AES, Hash) are successively applied on both dataset, and the actual Association Tests are perform by a third party (TPS).
PrivAS is the implementation of our previous work: https://ieeexplore.ieee.org/document/9119132
Introduction¶
Large samples of cases and controls need to be sequenced at the genome or exome level to perform association tests on rare variants. Clinical investigators often have easy access to the DNA of patients, but it is sometimes more difficult for them to recruit controls. Furthermore, they would be more inclined to use a bigger part of the budget on sequencing more patients than healthy controls, especially as more and more reference panel datasets are available.
Alas, those reference datasets are mostly aggregated data, only providing the frequencies of the variants among the individuals of the panel and not individual genotypes. The most optimal association tests need access to those individual genotypes to be performed. This sharing of sensible data represents a security risk for health data as well as intellectual property.
So we devised a method to allow two parties to pool their data together will compromising their privacy.
Security¶
Let us identify what information is required when performing association tests, and which parts of this information must remain private.
Basically for each genomic region or gene, we have to establish a list of variants for each party. Then we must pool those variants together and compare the genotypes repartition across both datasets.
We cannot disclose the individual genotypes of the variants, because they represent health data. However, the genotypes themselves are not sensible, as long as they are not linked to their variants or genes.
So by simply hashing the variants (chromosome / position / allele) and gene name, it is perfectly possible to perform anonymous association tests, where we can measure a signal without knowing which actual variants and genes are involved.
This test could be performed by one of the two parties. However, since this party is privy to the hashing key it is straightforward for them to reconstruct the complete information of the other party. So neither party will perform actual computations, that will be deported on an independent third party server.
In our implementation, the Reference Panel Provider (RPP), whose main role is to provide access to datasets of control genotypes, acts as a proxy between the Client (who possesses genotypes data for patients) and the Third-Party Server (TPS).
This obviously raises security concerns, as RPP could easily intercept data transiting between the Client and TPS. To prevent this risk, symmetrical encryption between Client and TPS is used, through an AES encryption key, unknown to the RPP. The choice of this technology (instead of the asymmetrical encryption proposed by RSA) is guided by the size of encrypted data. Indeed, genetic data are very voluminous and the RSA encryption overhead would lead the very big messages, so these exchanges are protected by AES.
Once again, the Client and TPS have to agree on a unique AES encryption key, with RPP acting as a proxy and able to intercept this key. Here, the Client generates a unique AES key for the current working session, and sends it to TPS through RPP after encryption with the asymmetrical RSA technology. The Client uses TPS’s RSA Public Key to encrypt the AES Key, and only TPS can decrypt it, by using its own RSA Private Key.
At last, to prevent eavesdropping on the networks, all messages between RPP and the Client are protected using an RSA Key pair generated by the Client, and unique to the current working session. The security layer is similar to the SSL encryption used on https servers.
The complete data workflow is summarize in the following graph.
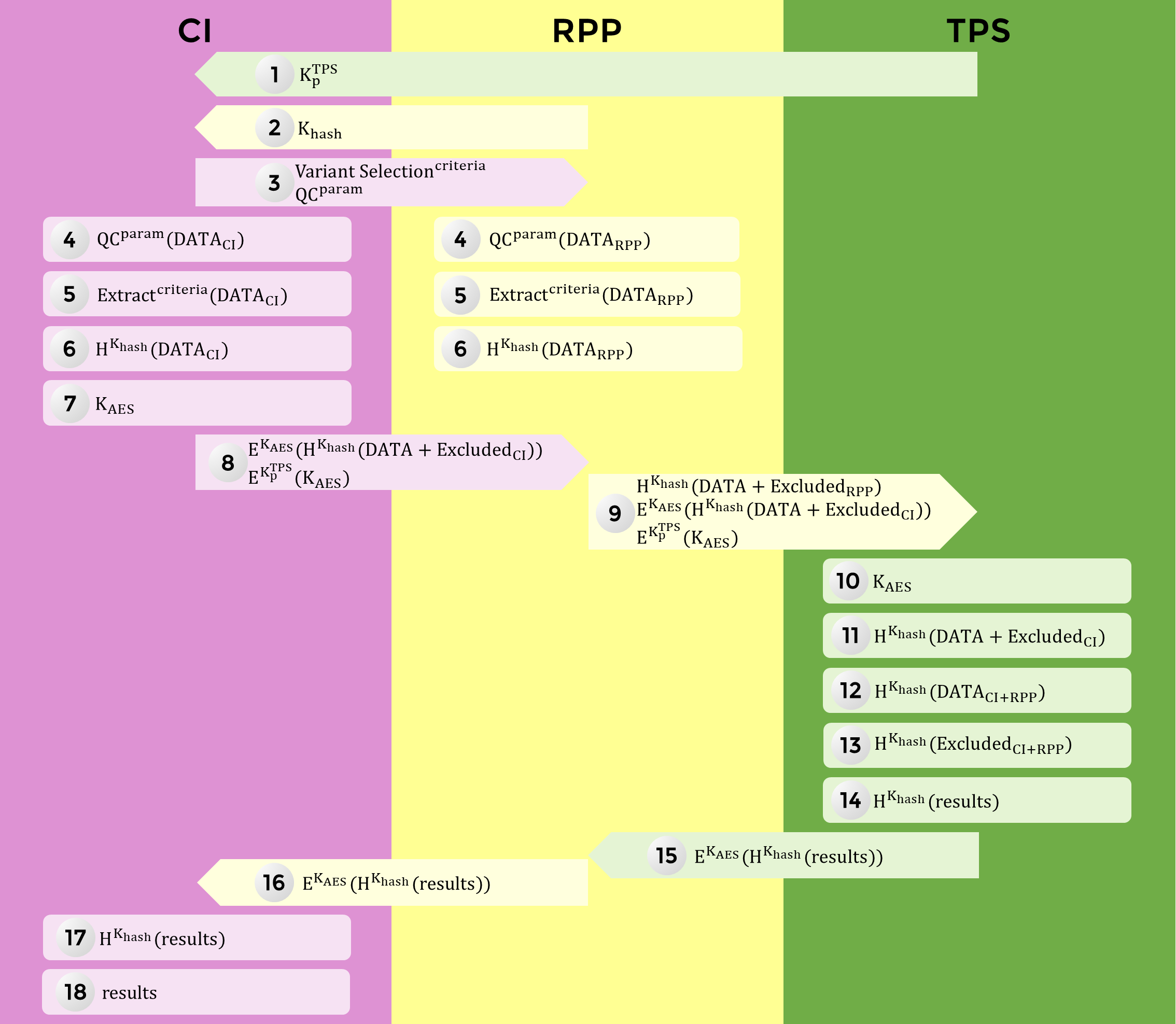
Client gets TPS’s
RSA Public KeyClient gets the
Hashing Keygenerated by RPP for this sessionClient sends the Variants
Selection Criteriaand theQC parametersto RPPClient and RPP perform the QC on their data and produce a list of
Excluded variantsClient and RPP extract variants from their data, according to the
selection criteriaClient and RPP use the
Hashing Keyto hash variants and gene names,Client generates an
AES Keyfor this sessionClient sends to RPP: the AES-encrypted, hashed
ClientData/ExcludedClientVariantsand the RSA-encryptedAES KeyRPP sends to TPS: the hashed
RPPData/ExcludedRPPVariants, the AES-encrypted hashedClientData/ExcludedRPPVariantsand the RSA encryptedAES KeyTPS uses its
RSA Private Keyto retrieve theAES KeyTPS uses the
AES Keyto retrieve the hashedClientData/ExcludedClientVariantsTPS pools the hashed
ExcludedRPPVariantsand hashedExcludedClientVariantsto produce a list of excluded variantsTPS pools the hashed
ClientDataand hashedRPPData, and performs Associations TestsTPS gets as
resultsp-values for hashedgenenamesTPS sends AES-encrypted hashed
resultsto RPPRPP relays those AES-encrypted hashed
resultsto ClientClient uses the
AES Keyto retrieve hashedresultsClient reverts the hashing of the
genenames for clear textresults
On the importance of Quality Control¶
In the context of PrivAS, association tests are performed on datasets from different organisations. Those data were produces at different times, on different sites, sometimes using different technologies. Under those conditions, a strong batch effect has to be expected. Only a careful selection of variants and regions through a thorough quality control (QC) process can help lower this effect to a minimum.
In our team, we developed such a QC, relying on various metrics. See https://gitlab.com/gmarenne/ravaq for further explanations.
In PrivAS, prior to the extraction of the variants of interests, a QC is always performed on both sets. The fine-tuning of the QC parameters is under the Client supervision, but the default values should be adequate for most datasets.
When a variant is present in one dataset but not in the other, it has a great weight on the results of the association tests. So, it is important to understand why the variant has not been found in the second dataset. There are basically 3 possibilities:
the
variantallele is not present in any of the individuals from the datasetthe position of the
variantwas badly or not covered at all during the sequencing of this set. That is why, along with thegenotypes, both parties also send abed fileof regions that were satisfyingly covered during the sequencing. Only positions found in the intersection of the beds from both parties are included in the tests.although well covered, the
variantwas removed during the QC. That is why both parties also send a list ofvariantsthat were excluded during the QC. Avariantfiltered during the QC by any party is also completely removed from the tests. PrivAS ensures that the same QC parameters are used by every party.
Performances¶
Since only the variants’ positions and gene names are hashed, the performances of the WSS Association Tests are the same as the tests on clear text data.
Evolutions¶
At this time, only the WSS Burden test is available in PrivAS, but we are currently implementing more tests (such as SKAT).
Availability¶
PrivAS can be downloaded from GitHub : https://github.com/ThomasLudwig/PrivAS
PrivAS is distributed as 3 java modules (one for each party involved). If you don’t wish to provide a Reference Panel, only the Client module is required.
Read more about:
the Client module
the RPP module
the TPS module
The server alanine.univ-brest.fr port 6666 provides data from the FrEx reference Panel (http://lysine.univ-brest.fr/frexac/). FrEx data contain exome+UTR sequencing of 574 a priori healthy individuals, recruited around 6 major cities in France (Bordeaux, Brest, Dijon, Lille, Nantes and Rouen). The Third Party Server TPS associated to this RPP is the datarmor supercomputer from ifremer (https://wwz.ifremer.fr/Recherche/Infrastructures-de-recherche/Infrastructures-numeriques/Pole-de-Calcul-et-de-Donnees-pour-la-Mer).
Contact¶
For any question about PrivAS, or for help in setting up RPP/TPS servers, you can contact me at ludwig@univ-brest.fr .